Visualize your fitted
non-linear dimension reduction model
in the high-dimensional data space
Joint work with Prof Dianne Cook, Dr Paul Harrison, Dr Michael Lydeamore, Dr Thiyanga S. Talagala
Motivation
Single-cell gene expression: same data, different NLDR + hyper-parameters
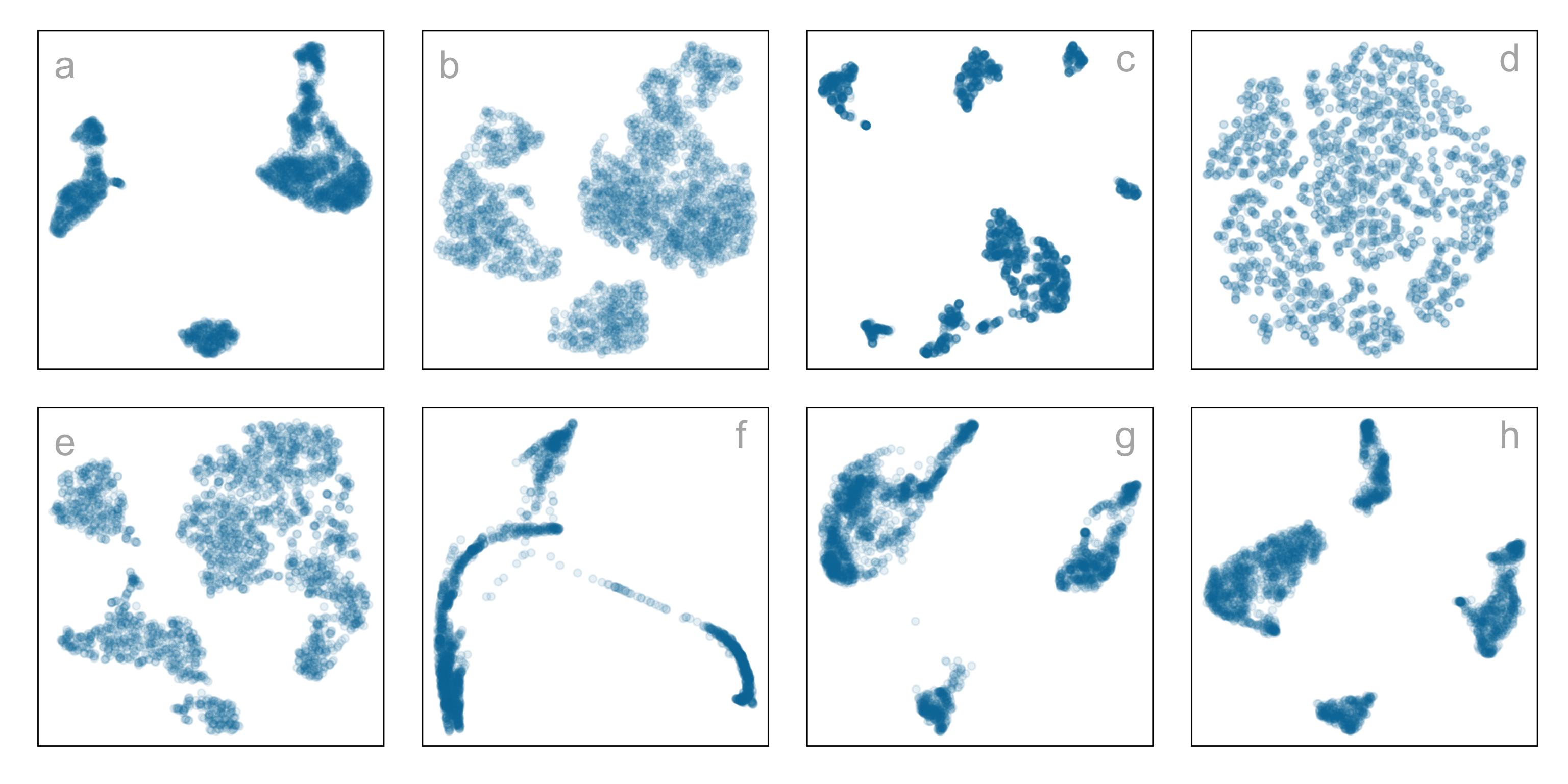
Which is the most reasonable representation of the structure(s) present in the
high-dimensional data?
How do you decide which is the most reasonable representation?
This is the published figure.
Peripheral Blood Mononuclear Cells (PBMC)
Here is the \(9\text{-}D\) data viewed using a grand tour, linear projections into \(2\text{-}D\).
Software: langevitour
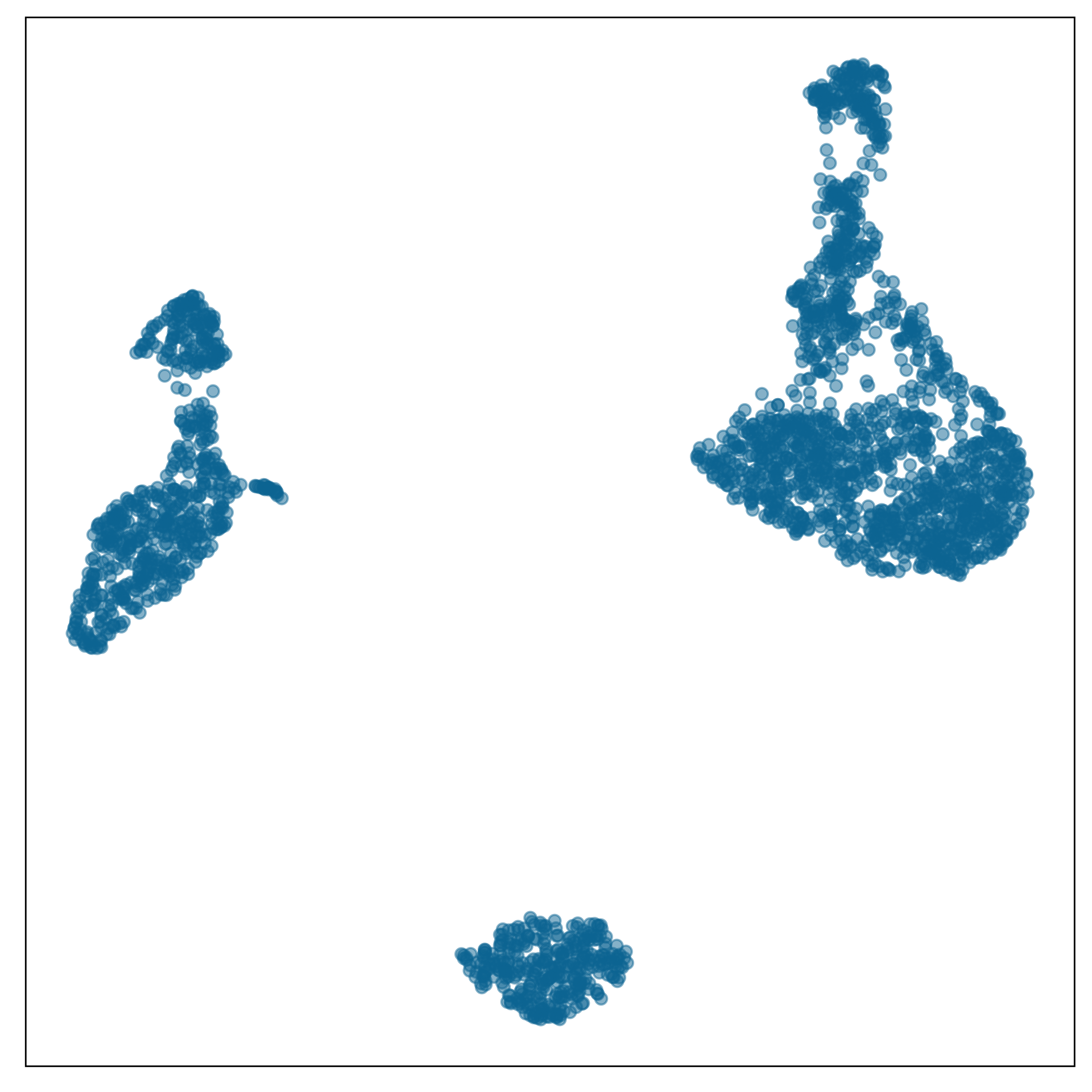
Show “model-in-the-data-space”
data-in-the-model-space
What is the model?
data-in-the-model-space
model-in-the-data-space
Overview of method
1. Construct the \(2\text{-}D\) model
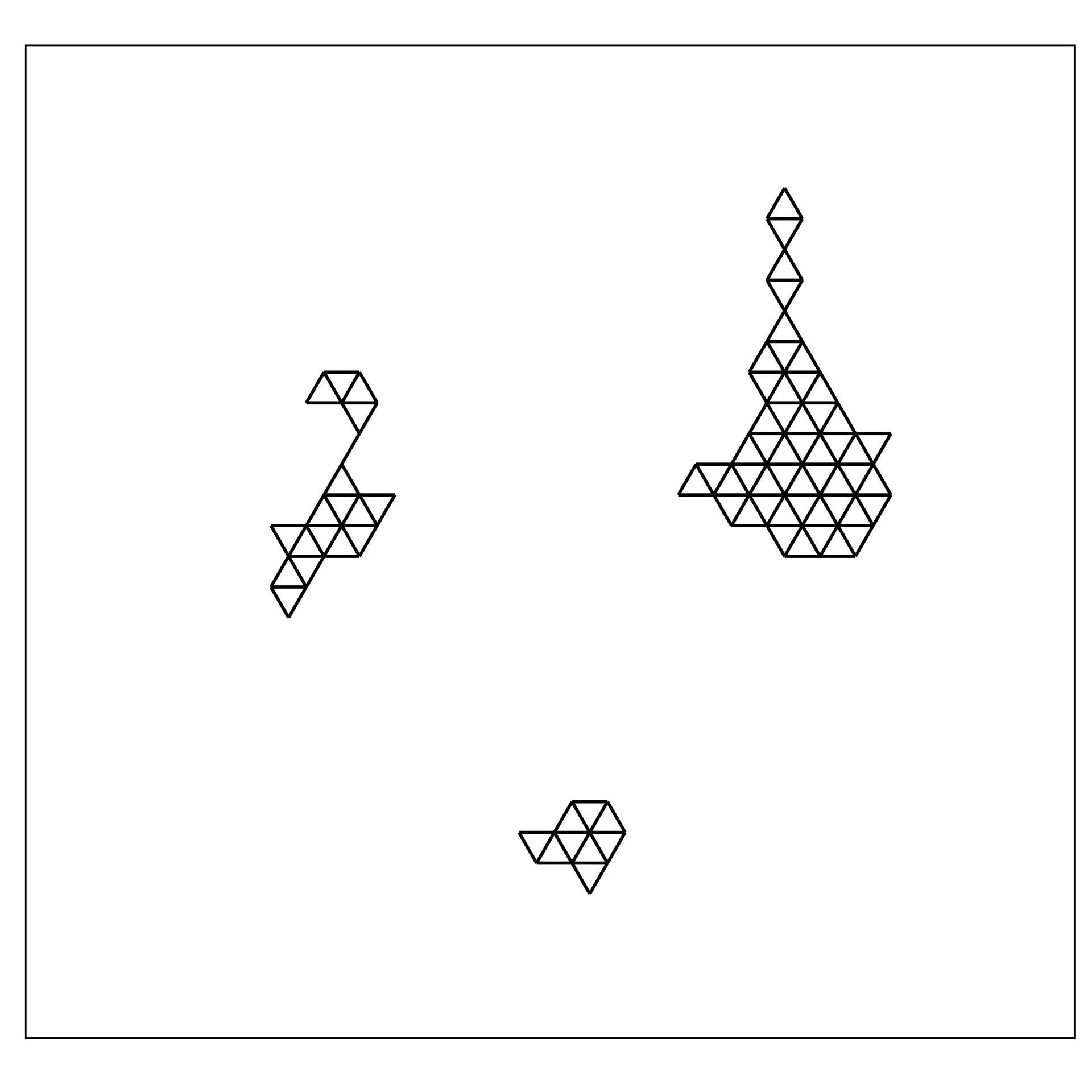
2. Lift the model into high-dimensions
Steps of the algorithm
1. Construct the \(2\text{-}D\) model
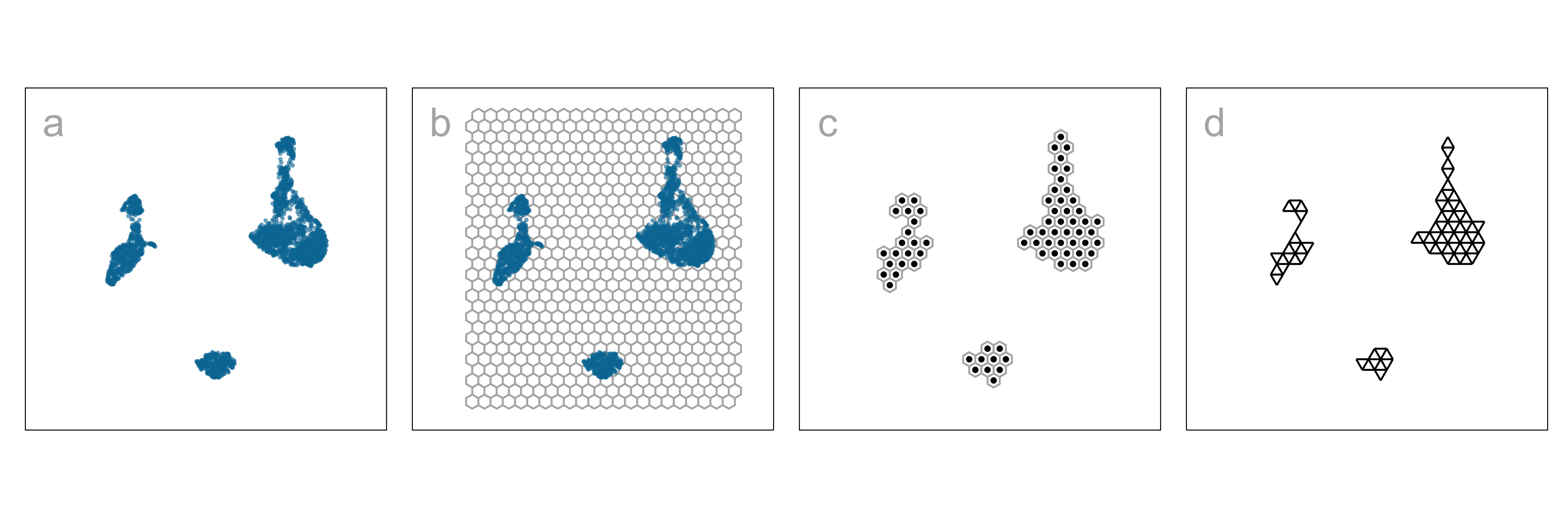
- NLDR layout, b. hexagon bins (
hex_binning()andgeom_hexgrid()), c. bin centroids (extract_hexbin_centroids()), d. triangulated centroids (tri_bin_centroids(),gen_edges()andgeom_trimesh()).
Steps of the algorithm
2. Lift the model into high-dimensions
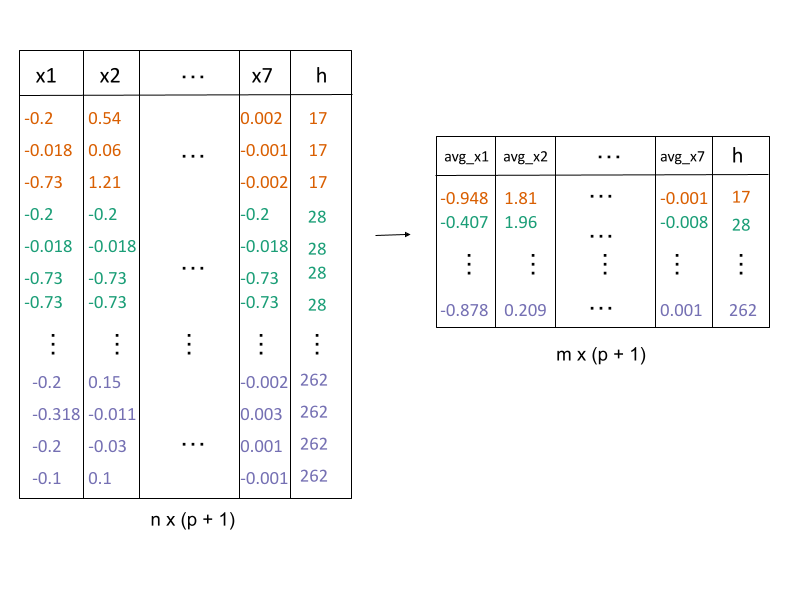
avg_highd_data()
show_langevitour()
RMSE of candidates
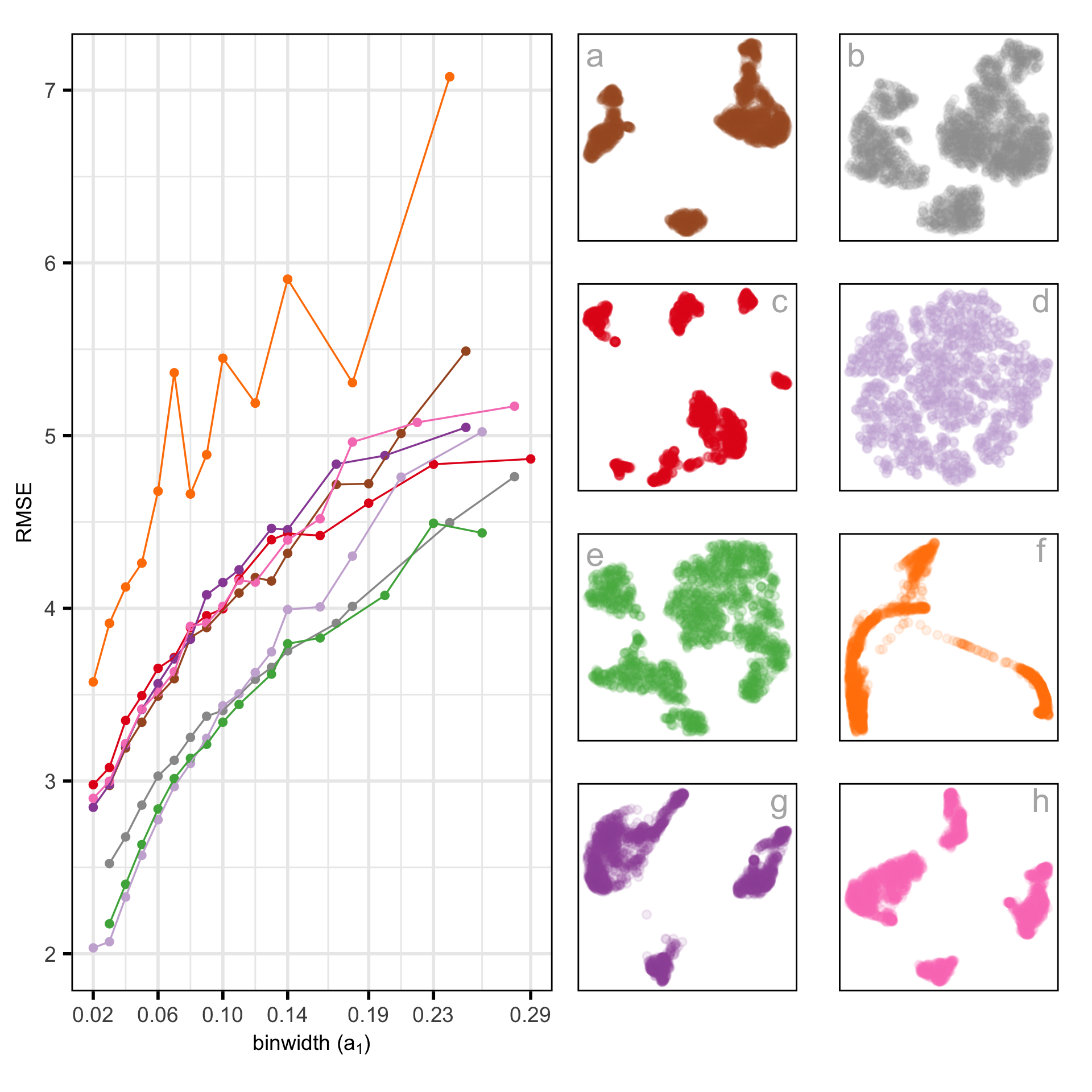
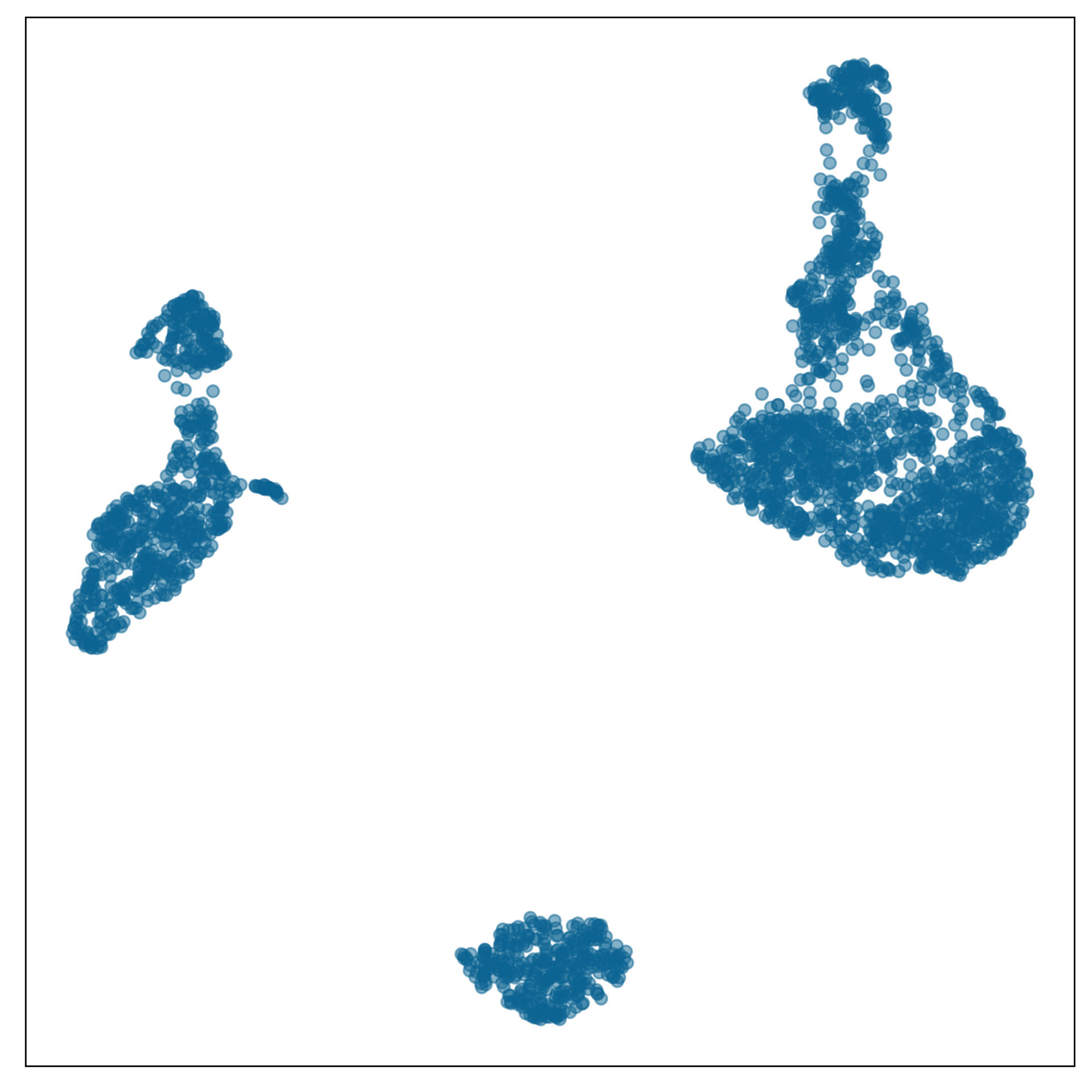
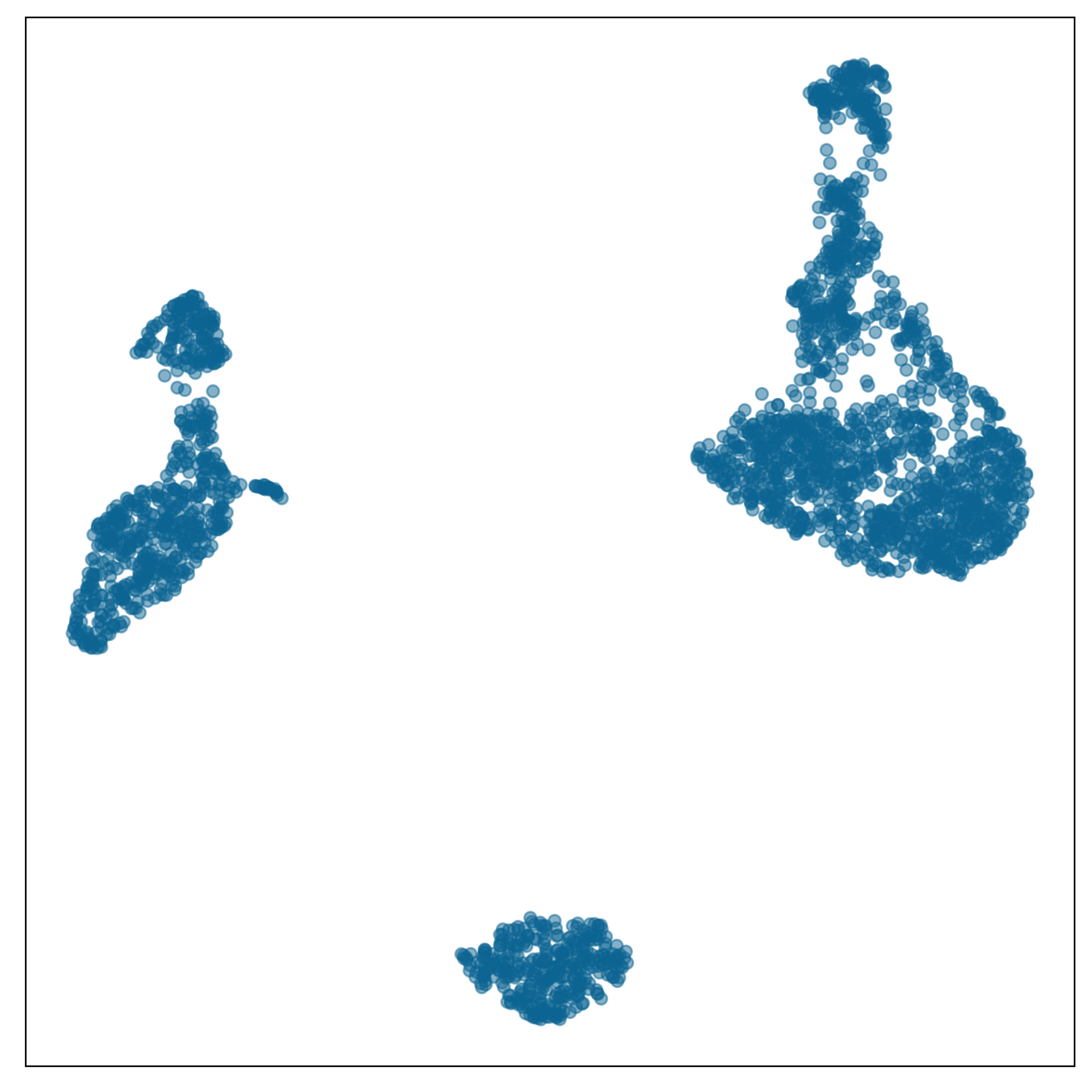
Chosen fit for PBMC data set
tSNE with perplexity: 30
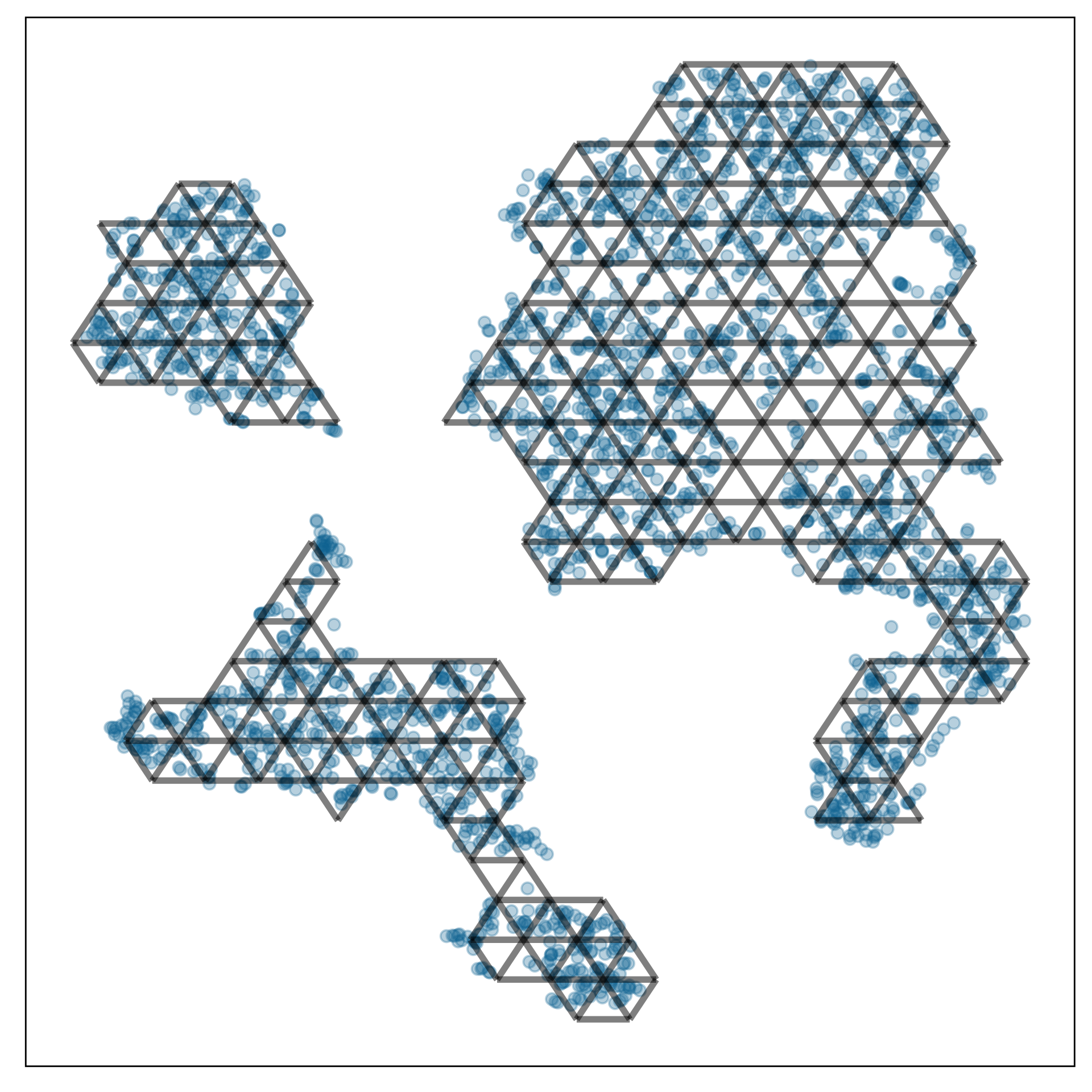
Clusters with small separations, non-linear clusters
Densed points, filled out clusters


quollr
questioning how a high-dimensional object looks in low-dimensions using r
R package

Draft paper
Jayani P.G. Lakshika 
Collaborators: Prof Dianne Cook, Dr Paul Harrison, Dr Michael Lydeamore, Dr Thiyanga S. Talagala